Almost everyone in the BioBricks realm seems to use a standard method for modifying their organisms: chemical transformation. Yet there is another method which is very promising.
In chemical transformation [3], some standard bacteria is grown, purified, mixed with some chemicals which cut open the bacteria, the new DNA plasmid is added to create some modified bacteria, the new DNA plasmid flows through the cut into the bacteria, everything is mixed with some more chemicals, allowed to heal & grow, and purified. (My naive translation of the process)
Note all the chemicals used? Chemicals can be expensive. And the amount of modified bacteria which results from this process is pretty low.
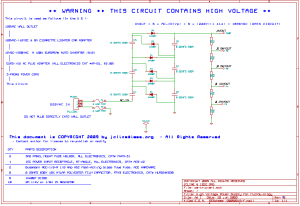
There’s an alternative method used more for yeast than bacteria: voltage-based transformation, electroporation [1]. With electroporation, some standard bacteria is grown, mixed with some simple chemicals, the new DNA plasmid is added, everything is given a quick high voltage zap (like lightening) which cuts open the bacteria, the new DNA plasmid flows through the cut into the bacteria, the bacteria is allowed to heal & grow, and purified. (Again, my naive translation of the process)
This eliminates some chemicals, although the process still requires some specialized equipment which can be troublesome (and expensive) — the voltage can be as high as 5 kV at 20 mA. (As high as the internal components of a CRT television, which, if accidentally touched, can be easily fatal.)
There’s another method though, that I haven’t seen mentioned: sonic transformation, sonoporation [2]. In sonic transformation, some standard bacteria is grown, some chemicals are added, optionally producing small bubbles, the new DNA plasmid is added, everything is given a loud blast of ultrasound (for example, at 40 kHz) which cuts open the bacteria, the new DNA plasmid flows through the cut into the bacteria, the bacteria is allowed to heal & grow, and purified.

In the research quoted below, sonoporation has shown to be much more effective at modifying bacteria than either chemical transformation or electroporation; plus, this is done without the expensive chemicals necessary for chemical transformation and without high voltage equipment necessary for electroporation.
From [2]:
More→