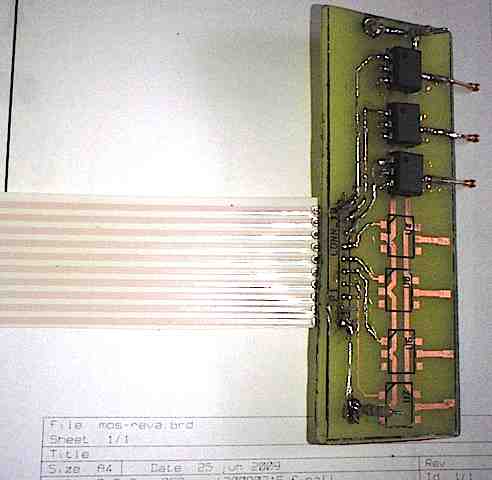
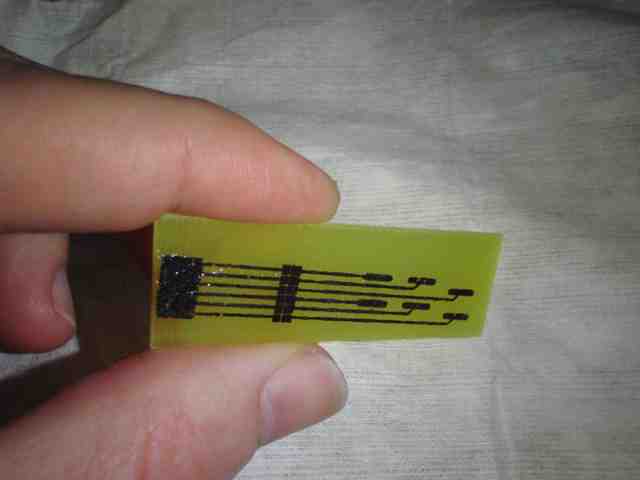
Imagine reading these kinds of instructions and performing such a task for a few hours: “Resuspend pelleted bacterial cells in 250 µl Buffer P1 and transfer to a micro-centrifuge tube. Ensure that RNase A has been added to Buffer P1. No cell clumps should be visible after resuspension of the pellet. If LyseBlue reagent has been added to Buffer P1, vigorously shake the buffer bottle to ensure LyseBlue particles are completely dissolved. The bacteria should be resuspended completely by vortexing or pipetting up and down until no cell clumps remain. Add 250 µl Buffer P2 and mix thoroughly by inverting the tube 4–6 times. Mix gently by inverting the tube. Do not vortex, as this will result in…” (The protocol examples used here are from Qiagen’s Miniprep kit, QIAPrep.)

Wait a minute! Isn’t that what robots are for? Unfortunately, programming a bioscience robot to do a task might take half a day or a full day (or more, if it hasn’t been calibrated recently, or needs some equipment moved around). If this task has to be performed 100 or 10,000 times then it is a good idea to use a robot. If it only has to be done twice or 10 times, it may be more trouble than it’s worth. Is there a middle ground here?
If regular English-language biology protocols could be fed directly into a machine, and the machine could learn what to do on it’s own, wouldn’t that be great? What if these biology protocols could be downloaded from the web, from a site like protocol-online.org ? It’s possible! (Within the limited range of tasks that are required in a biology lab, and the limited range of language expected in a biology protocol.)
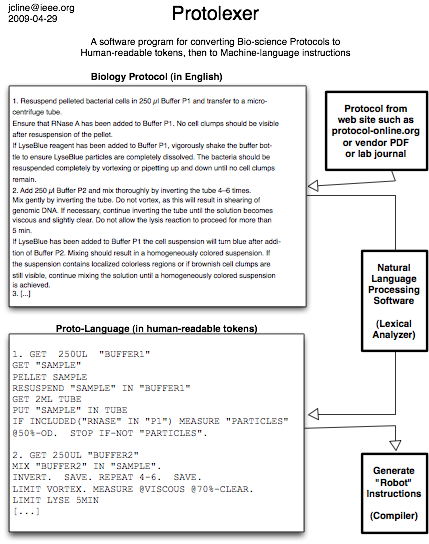
Biology Protocol Lexical Analyzer converts biology protocols to machine code for a robot or microfluidic system to carry out
The point of this prototype project is this: there are thousands of biology protocols in existence, and biologists won’t quickly transition to learning enough engineering to write automated language themselves (and it is also more effort than should be necessary to use a “easy-to-use GUI” for training a robot). The computer itself should be used to bridge the language gap. Microfluidics automation platforms (Lab on Chip) may be able to carry out the bulk of busy work without excessive “training” required.
More→