Imagine reading these kinds of instructions and performing such a task for a few hours: “Resuspend pelleted bacterial cells in 250 µl Buffer P1 and transfer to a micro-centrifuge tube. Ensure that RNase A has been added to Buffer P1. No cell clumps should be visible after resuspension of the pellet. If LyseBlue reagent has been added to Buffer P1, vigorously shake the buffer bottle to ensure LyseBlue particles are completely dissolved. The bacteria should be resuspended completely by vortexing or pipetting up and down until no cell clumps remain. Add 250 µl Buffer P2 and mix thoroughly by inverting the tube 4–6 times. Mix gently by inverting the tube. Do not vortex, as this will result in…” (The protocol examples used here are from Qiagen’s Miniprep kit, QIAPrep.)
Wait a minute! Isn’t that what robots are for? Unfortunately, programming a bioscience robot to do a task might take half a day or a full day (or more, if it hasn’t been calibrated recently, or needs some equipment moved around). If this task has to be performed 100 or 10,000 times then it is a good idea to use a robot. If it only has to be done twice or 10 times, it may be more trouble than it’s worth. Is there a middle ground here?
If regular English-language biology protocols could be fed directly into a machine, and the machine could learn what to do on it’s own, wouldn’t that be great? What if these biology protocols could be downloaded from the web, from a site like protocol-online.org ? It’s possible! (Within the limited range of tasks that are required in a biology lab, and the limited range of language expected in a biology protocol.)
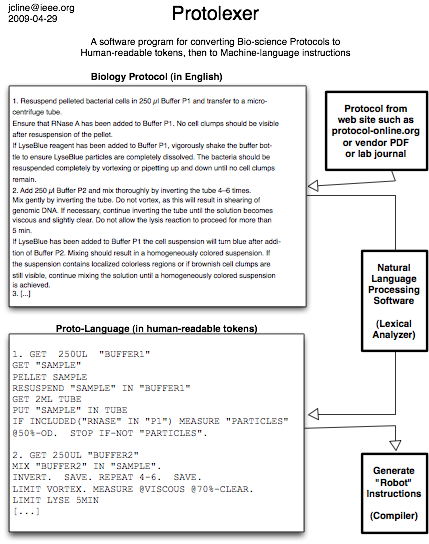
Biology Protocol Lexical Analyzer converts biology protocols to machine code for a robot or microfluidic system to carry out
The point of this prototype project is this: there are thousands of biology protocols in existence, and biologists won’t quickly transition to learning enough engineering to write automated language themselves (and it is also more effort than should be necessary to use a “easy-to-use GUI” for training a robot). The computer itself should be used to bridge the language gap. Microfluidics automation platforms (Lab on Chip) may be able to carry out the bulk of busy work without excessive “training” required.
I haven’t seen publications of this type for a biology context. This is the realm of “natural language processing” and significant research in Computer Science has made it relatively straightforward to translate well-formatted English into computer-understandable instructions. One unexpected source of pain is this: many of the engineers and computer scientists out there want to help biologists and often they try to do this by teaching engineering or computer science to the biologist. Too time consuming! Biologists should do what biologists do best: that’s biology and biological design (especially synthetic biologists). The computer scientists should force the machines to work for the biologists, rather than expect the biologists to learn the machines.
There are thousands of biology protocols (methods) and nearly all labs customize each protocol to their liking. This is fine. Use today’s incredible computer power to translate all these protocols directly into machine instructions. Get rid of the enormous wasted effort required to “program and calibrate” the machines which are supposed to serve the biologists: have the machines learn biology.
Here is a way to skip training the robot and have the machine read the protocol to automate itself: use lexical analysis on well-formatted biology text. For this project, I called it “the Protolexer”.
In prototype stage, it works like this:
jonathan$ make miniprep # JC's Protolexer # Copyright 2009 Jonathan Cline 88proof.com All Rights Reserved Choose the protocol you wish to run now: (Insert fancy UI here) 1. Plasmid DNA Purification Using the QIAprep 8 Miniprep Kit 2. Plasmid DNA Purification Using the QIAprep Spin Miniprep Kit and 5 ml Collection Tubes 3. Plasmid DNA Purification Using the QIAprep Spin Miniprep Kit and a Vacuum Manifold 4. Plasmid DNA Purification Using the QIAprep 96 Turbo Miniprep Kit 5. Plasmid DNA Purification Using the QIAprep 8 Turbo Miniprep Kit 6. Plasmid DNA Purification Using the QIAprep Spin Miniprep Kit and a Microcentrifuge Select: 1 You choose to run the Plasmid DNA Purification Using the QIAprep 8 Miniprep Kit
Internally, the protolexer uses perl to parse the QIAGen text. In addition, it can “read” the text from other protocol-online.org miniprep protocols.
The proto-language is in flux, though for the curious, it looks like this (everything uppercase is machine instructions, anything lowercase is unparsed and thus ignored):
RESUSPEND<- PELLET<- "SAMPLE" 250UL "BUFFER1" TRANSFER-> ->WELL ; IF-INCLUDED "RNASE" ADD<- Buffer P1. IF-NOT "PARTICLES" @50%-OD after resuspension PELLET-> ; ADD-> 250UL "BUFFER2" MIX-> INVERT-> SAVE 4–6 REPEAT ; MIX-> INVERT-> SAVE ; LIMIT VORTEX-> result shearing genomic IF continue INVERT-> SAVE LIMIT @VISCOUS @70%-CLEAR ; LIMIT LYSE-> 5MIN ; ADD-> 350UL "BUFFERN3" MIX-> immediately INVERT-> SAVE 4–6 REPEAT ; To avoid localized precipitation MIX-> but immedi- ately after addition Buffer N3. @50%-CLEAR ; ....
Compare that to another miniprep protocol pulled from protocol-online.org:
jonathan$ make miniprep # JC's Protolexer # Copyright 2009 Jonathan Cline 88proof.com All Rights Reserved Choose the protocol you wish to run now: (Insert fancy UI here) 1. Alkaline Lysis Mini Plasmid Preps Select: 1 You choose to run the Alkaline Lysis Mini Plasmid Preps "SAMPLE" @50%-OD 1.5ML "LMM" or "TERRIFIC-BROTH" ADD-WITH 75ug/ml "AMP" MIX-> SAVE ->SAVE CENTRIFUGE-> "SAMPLE" 7-8K 2MIN ASPIRATE-> RESUSPEND<- 50UL 25mM Tris pH 8 10 mM EDTA; <-UNSEAL ADD-> 100UL 1% "SDS" 0.2M "NAOH" (5ML = 100UL 10M "NAOH" ADD<- 4.4ML "DDW" then 500UL 10% SDS) ; ADD-> forcefully you don't need VORTEX-> ADD-> 75UL "KOAC" VORTEX-> ADD-> 100UL "PHENOL-CHI3" SEAL-> VORTEX-> CENTRIFUGE-> 13000RPM 2MIN DRAIN-> ADD-> 500UL "ETHANOL" ; VORTEX-> CENTRIFUGE-> 13000RPM 5MIN ASPIRATE-> DRAIN-> "ETHANOL" RESUSPEND<- 50UL "TE" DIGEST-> 2-5UL ADD-> 1UL preboiled 10mg/ml "RNASE" preparation) FINISH
The source code for Protolexer is in perl. It’s relatively simple and clean. It’s a simple proof-of-concept, and hopefully a computer science programmer out there who knows LEX and YACC will dig into writing a full parser to machine code. The verbs are simple. The nouns are well defined. The formatting can be specified. It is a straightforward software project with some very cool ramifications.
