SynBioSS (Synthetic Biology Software Suite) is a suite of software for the modeling and simulation of synthetic genetic constructs. SynBioSS utilizes the registry of standard biological parts, a database of kinetic parameters, and both graphical and command-line interfaces to multiscale simulation algorithms. SynBioSS is available under the GNU General Public License. — Anthony D. Hill, Jonathan R. Tomshine, Emma M. B. Weeding, Vassilios Sotiropoulos, and Yiannis N. Kaznessis, Bioinformatics 2008 24(21):2551-2553; doi:10.1093/bioinformatics/btn468
Sounds neat, let’s try it. Interestingly, the iGEM participants and biologists, in discussions of modeling, have thrown their hands in the air & state that it is difficult or impossible to model biology. Maybe SynBioSS can do the impossible? Except: There is no specific installer available for OS/X (as of this writing) and it seems there are many assorted packages required.
Here are my install summary/notes/fixes for getting SynBioSS (version 1.0.1) running on OS/X (Leopard 10.5.5):
- Download synbioss, untar. I used synbioss 1.0.1 (latest as of now).
- Make sure python is installed; install with fink if necessary. I have python 2.5.
- Run it, see what it complains about (hopefully nothing, though probably something): /sw/bin/python2.5 synbioss.py My attempt complained about each of the following, in turn.
- Install python gtk libs using fink. fink install pygtk2-py25
Note because fink is installing this in it’s ‘fink location’ of /sw and is compatible with fink, then be sure to run python using fink’s python (/sw/bin/python2.5), not Apple’s built-in python. And since it’s using GTK, that means running X11.App on OS/X. - Install sympy manually (it is not included in fink), by downloading latest version. http://code.google.com/p/sympy/ This is a python library so no build required.
- Install Unum manually (it is not included in fink), by downloading latest version. http://sourceforge.net/projects/unum/ There seems to be a darwinport of this available, however I don’t think it works since the original Unum web site from 2004 seems broken. Anyway, I am not running darwinports, I use fink.
- Install libsbml manually (latest 3.2.0). http://sbml.org/Software/libSBML Requires port compilation using ./configure .
By default, libSBML only builds the C and C++ API library. If you
want to configure libSBML to build the Java, C#, Python, Perl, Lisp,
MATLAB, Ruby and/or Octave API libraries as well, add the flags
–with-java, –with-csharp, –with-python, –with-perl, –with-lisp,
–with-matlab, –with-ruby, and/or –with-octave to the configure
command.
[…]
Making libSBML accessible to other software on Unix, Linux and MacOS X systemsOnce the libSBML files are installed as described in the previous section, you may need to do one more configuration step so that you and your users’ software can find the libSBML library files at run time. On most Linux-based platforms, this must be done either by running the system program ldconfig as user ‘root’ (consult the man page for ldconfig if this is unfamiliar), or else each user must set the environment variable LD_LIBRARY_PATH in their command shells. (On MacOS X, the variable is named DYLD_LIBRARY_PATH. Also, there is no equivalent on MacOS X to the ldconfig facility.) The path added to the LD_LIBRARY_PATH or DYLD_LIBRARY_PATH variable must be the directory used as the value of the –prefix=DIR option used during the configure step discussed above.
For my configuration, I used:
$ export DYLD_LIBRARY_PATH=/usr/lib
$# ./configure –with-python=/sw –includedir=/sw/include –bindir=/sw/bin –with-matlab=/Applications/MATLAB_R2008a/
I tried to configure with java support, however it complained that java 1.4.0 was required, so forget it. Luckily I already have libxml installed (or maybe it is OS/X built-in); otherwise likely install with fink. Then make . Don’t make install without setting the proper path configuration to /sw because otherwise libsbml defaults to install in /usr/ which is not correct on OS/X. (Try make install and verify paths are correct.) If correct then sudo make install . If everything goes well, then install should show the important step, copying build/_libsbml.so -> /sw/lib/python2.5/site-packages. I ended up manually copying libsbml.dylib to /usr/lib manually, though I think this made things more messy on my system.. though symbioss found the library. Also, I ended up doing both the configure and make as root, to get around some strange problem with the dylib; the libsbml.so was pointing to “../libsbml.dylib” internally, instead of /usr/lib/libsbml.dylib. Odd.
- Install netcdf and pycdf-0.6.3b.
- Shake head slowly at developers who use non-integrated scripting languages which require so many external dependencies.
- Run X11.App and start a Terminal. Set the python environment (PYTHONPATH) to find the installed prerequisites. Run synbioss as follows.
- At this point, I received the following error: line 129 import _winreg Import Error: No module named _winreg . Some s/w is schitzo here, because I’m not running windows, and line 129 is checking for win32 o/s. I edited synbioss line 129 to use if 0: instead of if ‘win’ …
- Repeat the step to run synbioss again, and the X11-GTK-OS/X Synbioss screen appears:
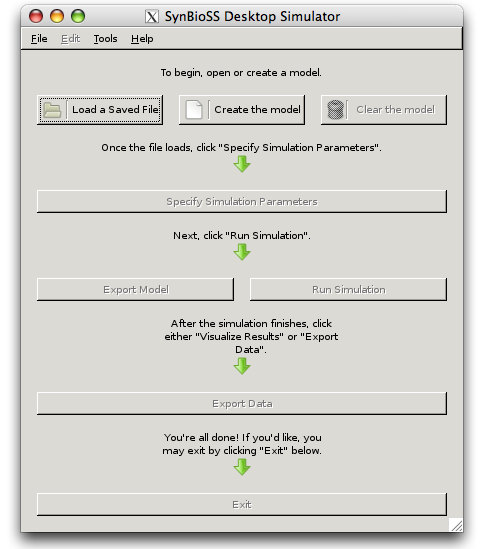
First is netcdf: export DYLD_LIBRARY_PATH=/usr/lib ; ./configure –prefix=/usr/local –disable-f77 –disable-f90 . Then make && make check. The tests from “check” should all pass. Then sudo make install . This should install stuff into /usr/lib . Now for pycdf: untar and run sudo /sw/bin/python2.5 setup.py install . This will copy stuff into /sw/ .
$ export PYTHONPATH=/Users/jonathan/local/sympy-0.6.2:/Users/jonathan/local/Unum-4.0
$ sw/bin/python2.5 synbioss.py
Note: Similar patch is required to filechoosergtk.py line 14. Other cases might exist.. fixed as I hit them (or could grep for them too)
Now that the simulator application is actually running, step back into the design phase to create something to simulate. Go to the SynBioSS Designer Web Interface and enter some BioBricks and such, in order to have an SBML file created from the BioBrick design:
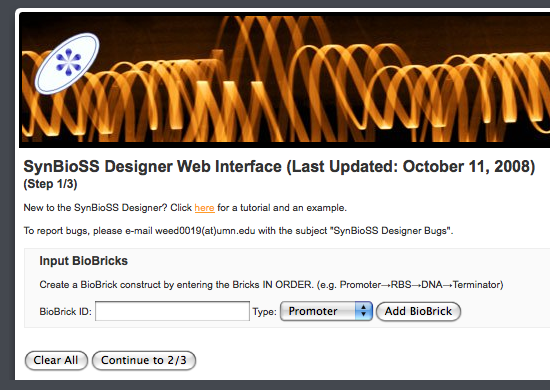
Unfortunately, the web interface isn’t so good at error checking — that’s a difficult job right now, and up to the designer to know what’s going on. So, my BioBrick device had errors (though I attempted to follow the documentation’s example), and the web interface reported an error “somewhere”. Looks like SynBioSS takes more than a day’s work to get running; so, look for a part 2…