My Part 1 of 5 review of the book, Beginning Perl for Bioinformatics, by James Tisdall, left off at Chapter 8, just before Tisdall explains associative arrays, gene expression, FASTA files, genomic databases, and restriction sites.
Tisdall: “For simplicity, let’s say you have the names for all the genes in the organism and a number for the expressed genes indicating the level of the expression in your experiment; the unexpressed genes have the number 0. Now let’s suppose you want to know if the genes were expressed, but not the expression levels, and you want to solve this programming problem using arrays. After all, you are somewhat familiar with arrays by this point. How do you proceed?”
Perl’s associative arrays are one of the most powerful aspects of the language. This is a good problem to examine using hashes. Solutions to this kind of problem in other languages (C or matlab) might create an N-dimensional array (or even NxM) as a matrix representation of the problem. In C, it might be solved using a lookup table possibly using a linked list, and the code to drive that needs to be written from scratch or borrowed from an external library. Perl has a built-in method to solve these kinds of problems.
The solution is to use a hash:
$gene_name = "triA";
$level = 10;
$expression_levels{$gene_name} = $level; # save 'level' on per-gene basis
This leads Tisdall to review biological transcription and translation, including code for DNA->RNA and RNA->protein data conversion. The code is given in long form and then optimized in further examples for speed using associative arrays. Recall the central dogma of biology:
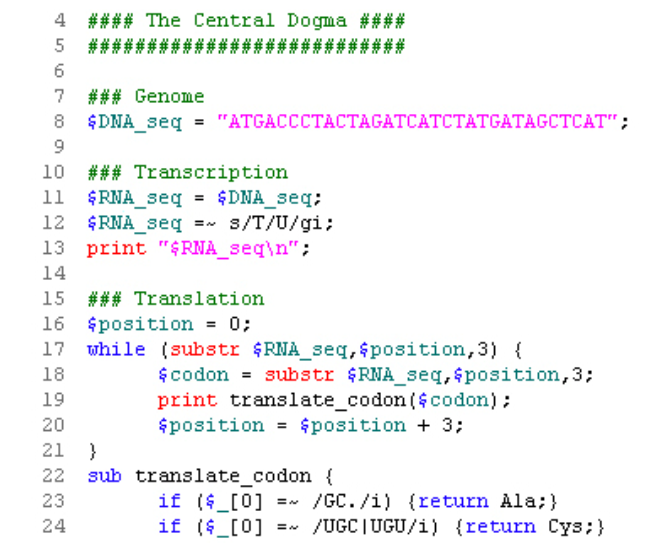
and so on and so forth.
Tisdall’s version is below.
# Tisdall Solution
#
# codon2aa
#
# A subroutine to translate a DNA 3-character codon to an amino acid
# Version 3, using hash lookup
sub codon2aa {
my($codon) = @_;
$codon = uc $codon;
my(%genetic_code) = (
'TCA' => 'S', # Serine
'TCC' => 'S', # Serine
'TCG' => 'S', # Serine
'TCT' => 'S', # Serine
'TTC' => 'F', # Phenylalanine
'TTT' => 'F', # Phenylalanine
'TTA' => 'L', # Leucine
'TTG' => 'L', # Leucine
'TAC' => 'Y', # Tyrosine
'TAT' => 'Y', # Tyrosine
'TAA' => '_', # Stop
'TAG' => '_', # Stop
'TGC' => 'C', # Cysteine
'TGT' => 'C', # Cysteine
'TGA' => '_', # Stop
'TGG' => 'W', # Tryptophan
'CTA' => 'L', # Leucine
'CTC' => 'L', # Leucine
'CTG' => 'L', # Leucine
'CTT' => 'L', # Leucine
'CCA' => 'P', # Proline
'CCC' => 'P', # Proline
'CCG' => 'P', # Proline
'CCT' => 'P', # Proline
'CAC' => 'H', # Histidine
'CAT' => 'H', # Histidine
'CAA' => 'Q', # Glutamine
'CAG' => 'Q', # Glutamine
'CGA' => 'R', # Arginine
'CGC' => 'R', # Arginine
'CGG' => 'R', # Arginine
'CGT' => 'R', # Arginine
'ATA' => 'I', # Isoleucine
'ATC' => 'I', # Isoleucine
'ATT' => 'I', # Isoleucine
'ATG' => 'M', # Methionine
'ACA' => 'T', # Threonine
'ACC' => 'T', # Threonine
'ACG' => 'T', # Threonine
'ACT' => 'T', # Threonine
'AAC' => 'N', # Asparagine
'AAT' => 'N', # Asparagine
'AAA' => 'K', # Lysine
'AAG' => 'K', # Lysine
'AGC' => 'S', # Serine
'AGT' => 'S', # Serine
'AGA' => 'R', # Arginine
'AGG' => 'R', # Arginine
'GTA' => 'V', # Valine
'GTC' => 'V', # Valine
'GTG' => 'V', # Valine
'GTT' => 'V', # Valine
'GCA' => 'A', # Alanine
'GCC' => 'A', # Alanine
'GCG' => 'A', # Alanine
'GCT' => 'A', # Alanine
'GAC' => 'D', # Aspartic Acid
'GAT' => 'D', # Aspartic Acid
'GAA' => 'E', # Glutamic Acid
'GAG' => 'E', # Glutamic Acid
'GGA' => 'G', # Glycine
'GGC' => 'G', # Glycine
'GGG' => 'G', # Glycine
'GGT' => 'G', # Glycine
);
if(exists $genetic_code{$codon}) {
return $genetic_code{$codon};
}else{
print STDERR "Bad codon \"$codon\"!!\n";
exit;
}
}
I don’t like that approach so much. I would prefer to use a __DATA__ segment to reference the table data and parse it into memory at runtime (especially since the author hints that the data table may change). Something like this:
# Solution by JonathanCline
#!/usr/bin/perl
Main(@ARGV);
exit;
sub codon2aa {
while (<DATA>) {
chomp;
last if /__END__/;
if (/^([\w_]+)\s([\w_]+)/) { $trans{$1} = $2; }
else { die "parse error $_"; }
}
if (defined $trans{$_[0]}) {
return $trans{$_[0]};
}
else {
warn "can't translate ".$_[0];
}
return;
}
sub Main {
codon2aa();
# Verify subroutine by printing all key/value pairs
for $key (keys %trans) {
print $key." = ".$trans{$key}."\n";
}
return;
}
__DATA__
TCA S # Serine
TCC S # Serine
TCG S # Serine
TCT S # Serine
TTC F # Phenylalanine
TTT F # Phenylalanine
TTA L # Leucine
TTG L # Leucine
TAC Y # Tyrosine
TAT Y # Tyrosine
TAA _ # Stop
TAG _ # Stop
TGC C # Cysteine
TGT C # Cysteine
TGA _ # Stop
TGG W # Tryptophan
CTA L # Leucine
CTC L # Leucine
CTG L # Leucine
CTT L # Leucine
CCA P # Proline
CCC P # Proline
CCG P # Proline
CCT P # Proline
CAC H # Histidine
CAT H # Histidine
CAA Q # Glutamine
CAG Q # Glutamine
CGA R # Arginine
CGC R # Arginine
CGG R # Arginine
CGT R # Arginine
ATA I # Isoleucine
ATC I # Isoleucine
ATT I # Isoleucine
ATG M # Methionine
ACA T # Threonine
ACC T # Threonine
ACG T # Threonine
ACT T # Threonine
AAC N # Asparagine
AAT N # Asparagine
AAA K # Lysine
AAG K # Lysine
AGC S # Serine
AGT S # Serine
AGA R # Arginine
AGG R # Arginine
GTA V # Valine
GTC V # Valine
GTG V # Valine
GTT V # Valine
GCA A # Alanine
GCC A # Alanine
GCG A # Alanine
GCT A # Alanine
GAC D # Aspartic Acid
GAT D # Aspartic Acid
GAA E # Glutamic Acid
GAG E # Glutamic Acid
GGA G # Glycine
GGC G # Glycine
GGG G # Glycine
GGT G # Glycine
__END__
In the above script, the code remains as code and the data remains segregated to a data section. The inclusion of data directly into a script as a separate section is another powerful feature of Perl. Most likely the author or others would eventually place the data section into an external file or external database. In that case, the structure of my subroutine remains the same, only the file handle changes, from the built-in name DATA to some arbitrary FILEIN.
It’s a good reminder that, in Perl, most everything can be done in multiple ways, depending on style or design. It’s useful to look at multiple solutions to Perl programming problems to see the different types of solutions available.
In the following sections, the author shows how to implement simple FASTA file handling, restriction mapping, GenBank, a Protein data bank, and BLAST.